COVID-19 Resources for the Amateur Epidemiologist
From Mars 2020, Argentina government put the whole country under quarantine. Nothing not happened in every other place in the world. War against COVID-19 started. Data scientists all over the country (and the world) entered the battlefield with their finest algorithms and visualization hoping to provide insights not perceived by the most famous epidemiologists in the world ;-). Any way, I was not the exception in all that visualization/modelization madness. I spent almost a moth learning and writing simple models and visualizations.
I have made a list of some of the most useful resources for having fun with epidemic modeling.
- A nice article from fivethirtyeight.com explaining Why It’s So Freaking Hard To Make A Good COVID-19 Model
- EPIDEMICS MODELS 101 A serie of posts on Medium about the basic for modeling epidemic spreads.
- Tim Churches serie of posts about modeling COVID-19 using R language. From simple compartment (SIR) to log-linear models
- An article from Cheng et al. where the extend a SEIR model for including other compartment such as Exposed, Dead and Quarantine.
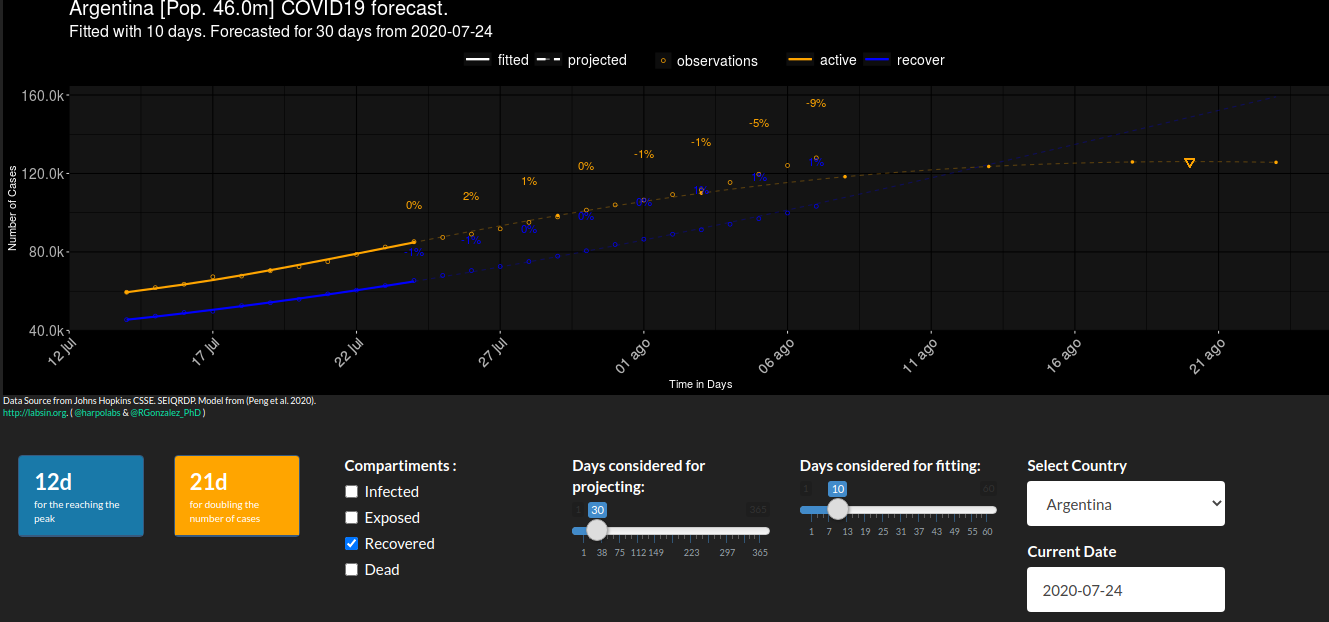
- My friend Rodralez from UTN Mendoza and me used Cheng et al. model for running our own simulations. You can find all our work here and here. In the repos you will find a Matlab and R implementation of the models, several visualizations and a simple shiny app..
- A dataset parsed from the official information provided by the Argentine Ministry of Health.
- Luis Mayorga and his team from IMBECU (CONICET ARGENTINA) developed a SEIR model for predicting the ICU beds occupied during the pandemic. They published an original research paper and provided a running model and a notebook for playing..
- Leandro Abraham from DHARMA and Eventbrite wrote a very nice notebook about predicting positive COVID19 patients using clinical information.
